In the year 2000, the first complete nuclear genome of a plant species – Arabidopsis thaliana – was released into the wild (a.k.a to bunch of salivating scientists). Less than twenty years later, we had a total of 1135 genomes… for Arabidopsis alone! Today we’re talking about Arabidopsis ‘races’, and how they are a powerful tool for unravelling plant secrets.
Arabidopsis likely diverged from its closest relative about 10 million years ago, and the invasive weed has since spread through Northern Eurasia. In the literally millions of years following, the species diversified into hundreds of race-like ecotypes: populations of plants that have settled in, and then adapted to, certain geological areas. While there has been some cross-talk (i.e, cross breeding) between the ecotypes, there has also been a whole lot of time spent alone – leading to evolution of traits that can differ from one ecotype to the next.
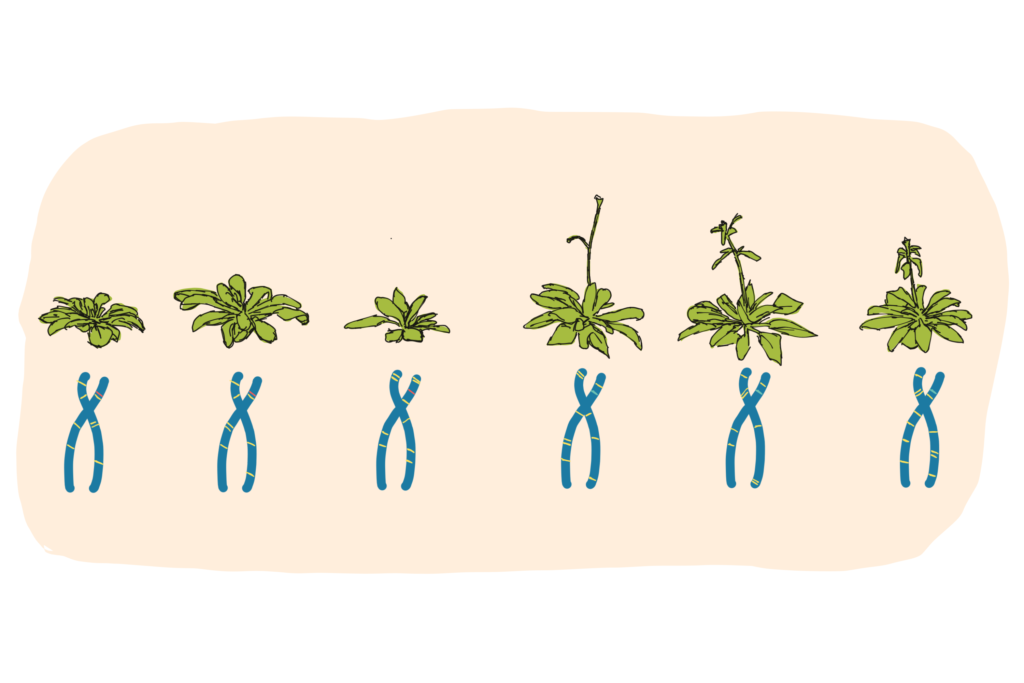
Arabidopsis ecotypes (also called accessions) can differ from each other in their size, shape, rate of growth, metabolic content, flowering behaviour, and response to light, stress, and diseases. In some cases, these variations can clearly be linked to the location of origin of the accessions, gifting scientists with vital information about environmental adaptation. For example, while some Arabidopsis accessions flower simply when they reach a certain age, and others take cues from day length, certain (Northern) accessions will only reproduce once they’ve experienced cold and can feel the spring again. This can be linked to a logical desire not to start flowering just as a harsh winter begins. (In the lab, we deal with this by shoving the poor plants in the fridge for a bit, and then bringing them out again).
Linking phenotype with environment is cool, but by obtaining the genomes of these ecotypes, we can take it one step further: looking for the DNA differences that cause the changes. Having access to many genotypes, belonging to many diverse ecotypes, makes it much easier to look for patterns of change that link ’cause’ (DNA) and ‘effect’ (phenotype).
For example, if two accessions are compared, one with an early flowering phenotype and one without, it can be tricky to discover which of the many differences in their two genomes facilitates flowering faster. However, if we now have 1135 accessions, and the 200 that flower early all have a certain change in a certain protein (in amongst the noise of all the other changes), but the non-early flowering ones don’t have this change…. well then we can start to make some functional links. It’s all very ‘A Beautiful Mind’.
Similarly, an early study investigating just 20 accessions (admittedly choosing a range of particularly diverse candidates), found that nearly 10% of known protein-coding genes were effectively ‘broken’ or missing in at least one accession. This kind of meta-data analysis can give clues about the importance of the missing genes in general (genes that are essential for life are more are likely to be maintained in all accessions), or perhaps their requirement under certain environmental conditions.
By the way, this in-species diversity is of course not unique to Arabidopsis. In crop plants, we tend to discuss cultivars- varieties that have been specifically breed for certain traits. Apart from the trait that has been bred (think, large size of fruits, leafy plants etc.), there are also extra variations that have come along for the ride. In tobacco, for example, some cultivars have a resistance response to the tobacco mosaic virus (TMV), which confines the bug to tiny sectors of the plant leaf. Meanwhile, in other cultivars, the virus spreads like wildfire, ultimately killing the plant. Some cultivars of tomato may be easily regenerated, allowing growth of entire tomato plants from tiny leaf pieces, while others refuse. And different cultivars of rice and wheat have various responses to abiotic and biotic stresses, including things like flooding, drought and salinty.
As we mention often on the blog, when it comes to nature, variety is the spice of life, which makes these ecotypes an immensely valuable resource to scientists. To hear about some particularly cool research, in which the morphological differences between ecotypes was exploited to better understand competition, check out our recent podcast release.
By the way, the most commonly used Arabidopsis accession, and the one who’s genome was sequenced first, is Col-0. This guy is a bit of a trickster, because it’s named for Columbia in the US, but is actually a central European accession. Other common players are Ler, Ws and C24.
You can read more about Arabidopsis ecotypes, and access publications specifically related to understanding accession diversity at the 1001 genomes project, here: https://1001genomes.org/