When you think of RNA, you might tend to think of a text-book perfect straight lines of As, Us, Cs and Gs. But the reality is, that within a living cell, RNA dances as a complex structure, full of twirls and loops. And when plants (and presumably other organisms – but this blog is about plants) are challenged with changing conditions, those structures begin to twist and change…
A recurring theme on this blog has been a focus on the amazing ways in which plants dynamically develop and react to changes, through complex cascades that tightly control the expression of their genes in response to stimuli.
You’re probably all familiar with the central notion of gene expression. That DNA, the stable inherited genetic material, is transcribed into transient mRNA, which in turn is used as a message that’s translated for the creation of working proteins. This straightforward process, DNA->RNA->Protein, is all well and good. But a need to tightly control, to keep responses in close check and balance, has resulted in the addition of layers and layers of regulation upon this basic scaffold.
An example of this regulation can be seen in the middle-man messenger RNA. Sure, mRNA is made (transcribed) from DNA, and used to make (translated into) proteins. But in between, it can also be capped (changed at the start of the RNA), polyadenylated (changed at the end of the RNA, by adding a bunch of As), spliced (cut up at different points in the middle and pasted together in different ways) and edited (have Cs changed to Us).
It can also be moved around – for example sequestered in blobs to keep it from being found for translation, or it can be permanently removed by degradation. And finally (at least for the point of today’s story), mRNA can undergo structural changes.
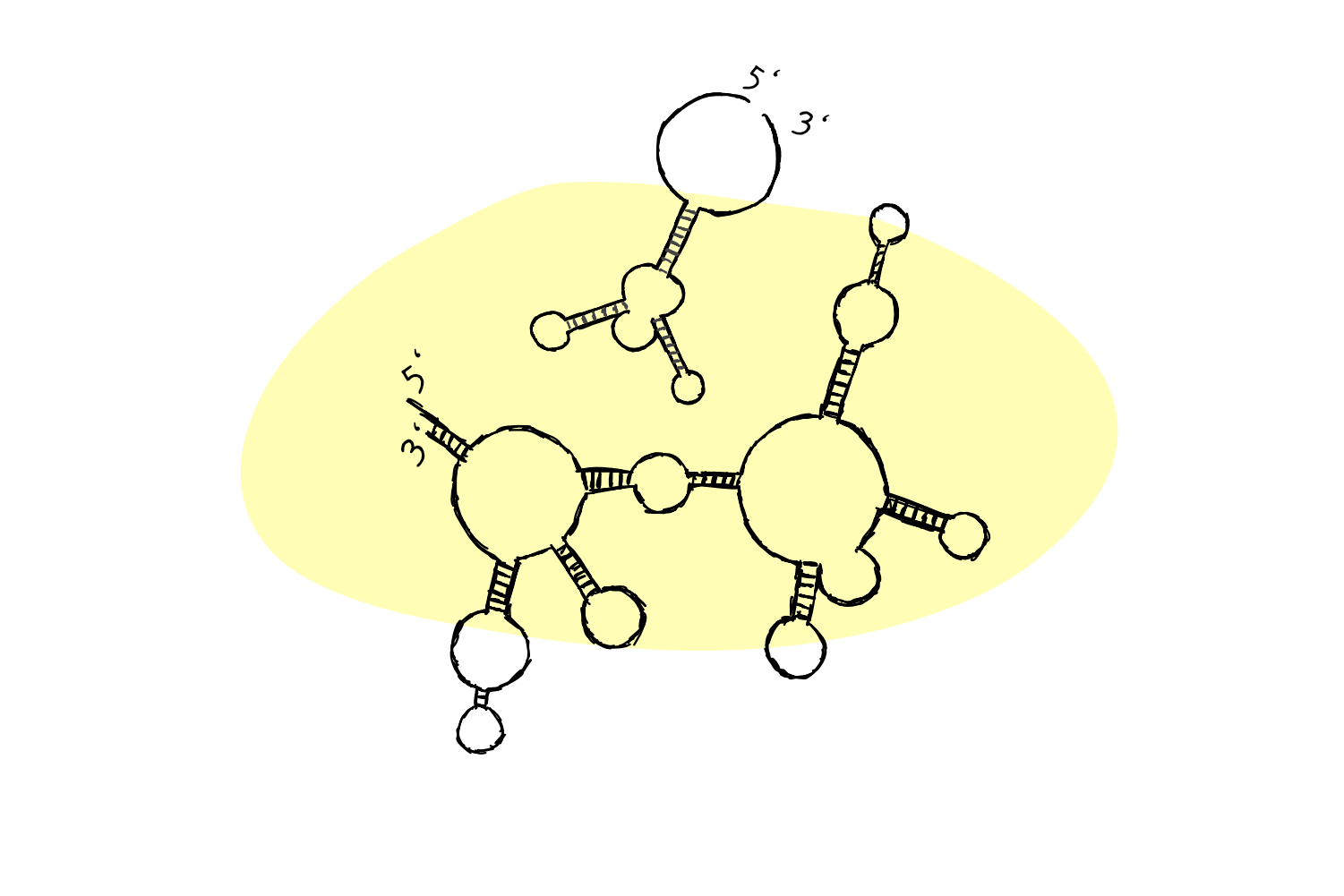
Because as it turns out, what we often imagine as this…

…might actually look more like this:

Note how a lot of the individual RNA bases have actually ‘paired up’ with other bases (A with U, C with G) to form these complex structures.
or maybe something like this:

This multi-dimensional physical structure of mRNA is actually a pretty important feature, that can have some pretty strong impacts on how well that mRNA in question can function. For example, having complex structure at their start (5’UTR), might make it more difficult for some mRNA to get translated into proteins by the cell’s translational apparatus. Similarly, structures at the end (3’UTR), might make it easier or harder for that mRNA to be recognised by the degradation machinery that seek it out for destruction!
Given the importance of these structures in defining gene expression, it’s not so surprising that several groups of scientists have worked to develop various methods to understand which structures exist, and how they dynamically change in response to stimuli.
More recently, techniques have become available to query mRNA structure in vivo and across the entire transcriptome. One of these methods, developed by Ding and colleagues for Arabidopsis in 2014, involves adding using a chemcial known as dimethyl sulfate (DMS) to cellular RNA. This DMS addition chemically modifies adenosine (A) or cytosine (C) residues that haven’t managed to pair up inside the twisted structure of the mRNA. When the mRNA is reverse transcribed – a process that makes cDNA that can then be sequenced – the modified bases block transcription, resulting in cDNA truncations. Truncations at different positions of the same mRNA molecule under different conditions, can then be used as an indication of different mRNA folding in those conditions.
Recently, David Tack and colleagues used this method to define how the RNA structurome (yep, that’s a word), changed in response to salt stress. They wanted to understand how mRNA structure changes, in order to better understand the implications for plant gene expression and adaptation to the stress. Salinity is a pretty major problem for crop productivity across the world, affecting nearly 1000 million hectares of agriculture fields and costing nearly 30 billion in crop losses annually!
Within plants, salinity causes cellular dehydration, disrupting ion homeostasis, promoting oxidative stress and reducing photosynthetic efficiency – to name just a few. Salt stress also leads to a whole lot of changes in gene expression: previous studies show massive changes in mRNA transcription and turnover, as well as changes in mRNA splicing patterns. But what about the structure of those mRNA?
Tack and colleagues grew arabidopsis seedlings until they were 24 days old, and then switched out the growth media with something a lot saltier (NaCl, 100 mM). And then they looked at the RNA structurome, focusing separately on changes that happened within the shoots, and those that happened in the roots of the plants.
As the authors discovered, there were changes to mRNA structures, but those changes seemed to differ depending on the precise region of the RNA observed. In both tissue types, the untranslated regions (UTR) at the start (5′) and end (3′) of the RNA, were more likely to change structure in response to salt. As it turns out, this might actually make a lot of biological sense – changes to these UTRs can have a big impact on how the RNA is translated, as well as how easily it’s targeted for degradation.
“These results highlight the potential of UTRs as structural mediators of regulation, consistent with their multiple functions” Tack et al., 2020, RNA

As you may have guessed from the fact that UTRs were more likely to change structure than the central coding sequence of the mRNA, another finding of the work was that different parts of the same RNA don’t always change in the same way… but also that they don’t always change in the same direction! The authors used the term ‘concordant’, if all three regions of the transcript (the 5’UTR, the coding sequence in the middle, and the 3’UTR) all changed structure in the same direction (all got more structure-y, or all got less).
As it turned out, RNA did NOT tend to behave concordantly.
Out of about 15,000 transcripts observed in the root, and another 15,000 or so in the shoots, only about 3,000 in each tissues were fully concordant (approx. 20%). Interestingly, these concordant RNA, when compared to their disconcordant brethren, also showed higher correlation between changes in mRNA structure and changes in mRNA abundance following salt stress.
In fact, the authors noticed that, globally, transcripts that tended to lose abundance under salt stress showed disproportionately increased mRNA reactivity (indicative of unfolding) under salt. Similarly, mRNA that increased in abundance was more likely to have decreased reactivity (increased structure) under salt. These findings led to the proposition: that stress-induced mRNA unfolding can contribute to decreases in mRNA abundance!
Finally, the scientists looked into which precise mRNAs were changing their structure and abundance, and found links to translation, photosynthesis, and – perhaps unsurprisingly – abiotic stress response.
All in all, the authors describe some interesting patterns of change in mRNA structure under salt stress, that putatively support the notion of salt-induced structural and (consequential) abundance changes to mRNA. This study is a fairly early player in the field of structuromics, and we’re interested to see how the idea of mRNA structural changes develops – as well as deeper into the complex questions of function, mechanism, and raison d’être!
References:
This blog post is based on the article:
Tissue-specific changes in the RNA structurome mediates salinity response in Arabidopsis. David C. Tack, Zhao Su, Yunqing Yu, Philip C. Bevilacqua, Sarah M. Assmann (2020), RNA.