Inside billions of cells all around us, DNA quietly makes messenger RNA (mRNA), which in turn acts as the template for protein synthesis.
In this equation, mRNA is the boring middle man. Unlike DNA, its lifetime is short, and it doesn’t get passed to the next generation. It also doesn’t get the protein life- catalyzing metabolic reactions, capturing the light from the sun, or transporting products across cell membranes.
Sometimes in our lives, we need a thrilling tale of an ordinary guy making a break for freedom….
Being a cell within a multi-cellular organisms is a lot like being many relationships at the same time. Being in a relationship can be rewarding, but requires a whole lot of work. And of course, great communication is key. Because different cells are good at doing or providing different things, the cells need to communicate to eachother what they need and when they need it. Think of toes demanding oxygen from lungs, or roots politely asking for energy from photosynthesizing leaves.
To stretch the metaphor to breaking point, while communication in short distance relationships is easy (tunnels through plant cell walls and membranes known as plasmodesmata allow movement of smallish molecules, ions and water from cell to cell), going long distance can be tricky.
Vascular plants use an old-school messaging system known as the vascular system to relay important goods and information through the plant. This system is roughly divided into xylem, mostly in charge of moving water from the roots to the leaves, and phloem, mostly in charge of moving sugars made in the leaves back down to other parts of the plant. In reality, it not just sugar and water, but a whole soup of things, including phytohormones, and signalling waves of calcium and reactive oxygen species to name a few.
And along for the ride, it turns out, is the greatest messenger of all: messenger RNA.
Scientists noticed it first in the phloem.
Phloem tissue is made of two cell types: companion and sieve. The companion cells are living, and are required to control and load the sieve cells, which are hollow and dead. Sieve cells have no nucleus and no DNA, meaning they can’t make their own RNA. But scientists found it, time and time again. Hundreds to thousands of different mRNA swimming though the sugar-saturated sieve straws.
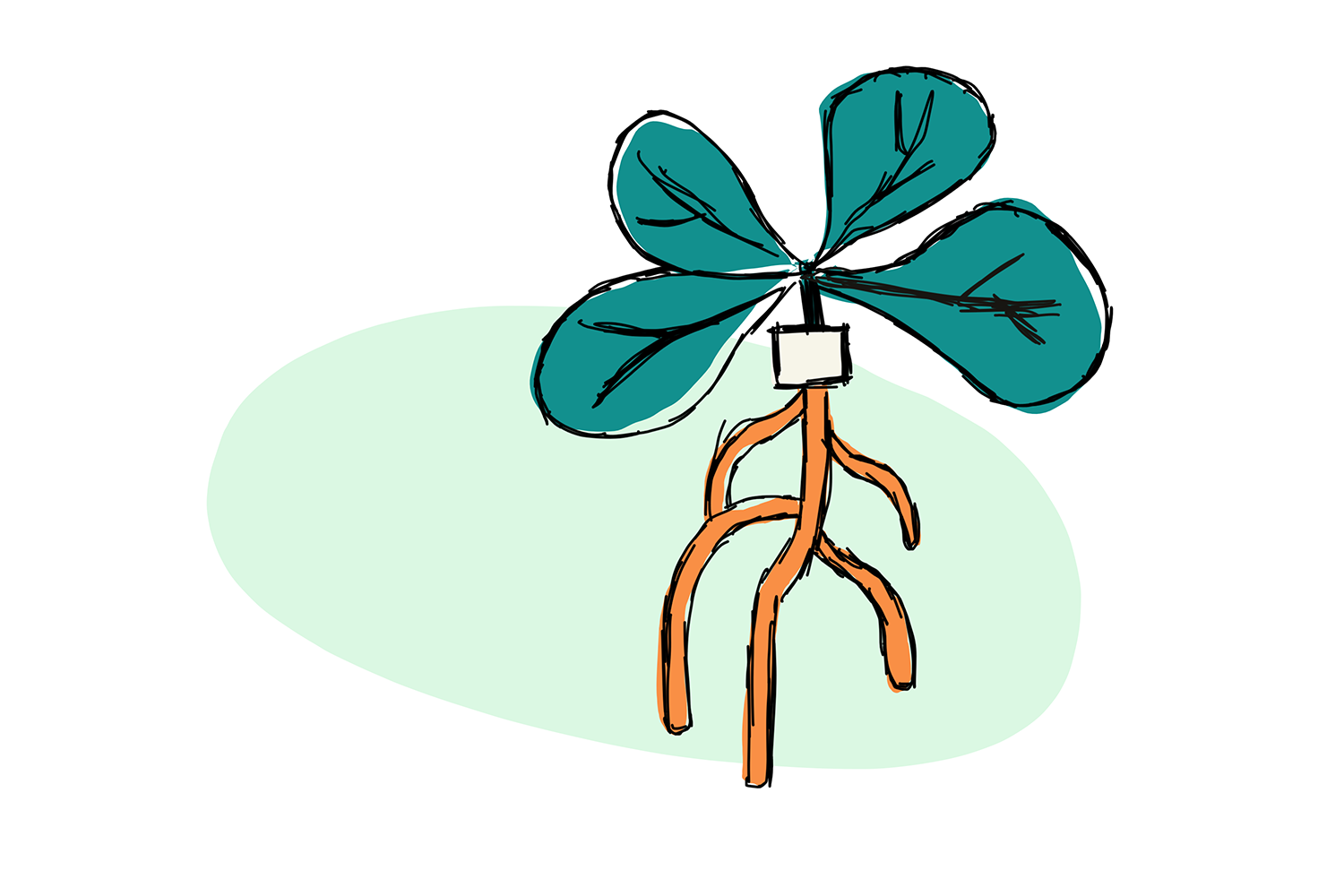
Scientists noticed that parasitic plants – which feed like vampires on the phloem-juice of their hosts, also acquired some of that host’s mRNA. More recently, a group made a graft of two different ecotypes or variants of the model Arabidopsis, putting together the root from one cultivar (e.g. Pedriza-0) and the shoot of another (Columbia-0) (Check out the Figure, below). After letting the plant grow together for a bit, they analyzed all the RNA in the top part, and all the RNA in the bottom part. Small differences in the genetic code of the two ecotypes, known as single nucleotide polymorphisms or SNPs, allowed them to see if the RNA belonged to the original plant, or had moved from its graft partner.
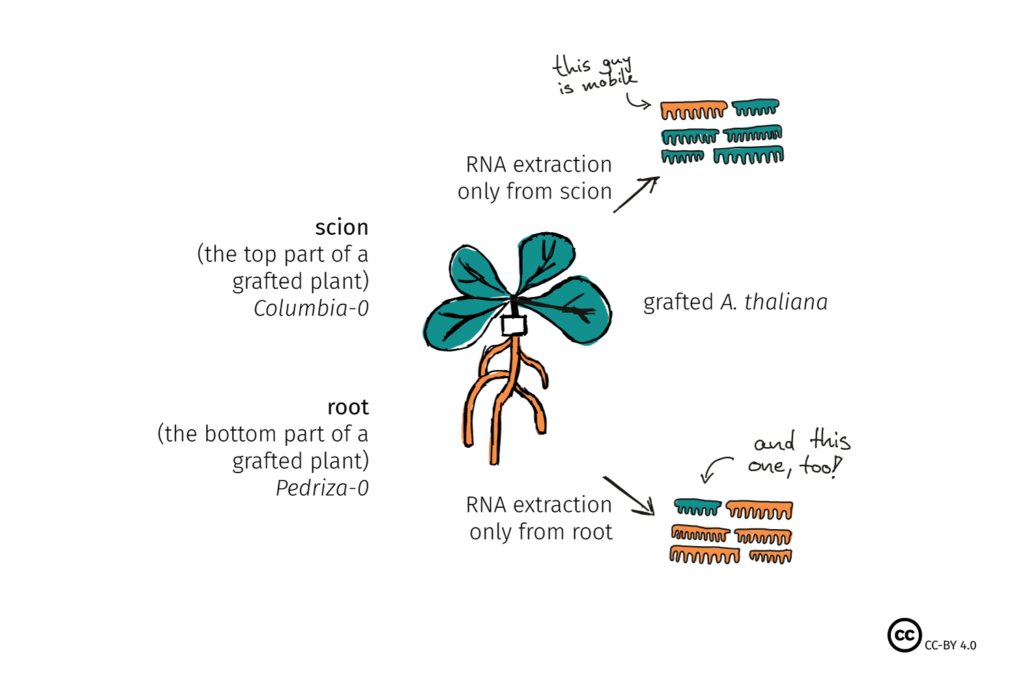
These experiments demonstrated that mRNA transfer can happen over long distance, across species. Furthermore, it seemed that some of the RNA was moving from the root of the plant to the shoot, against the natural flow of sugars in the phloem, suggesting that it might be moving ‘deliberately’. Add to that the phloem seemed to have a particularly large amount of stress-response and homeostasis-related mRNA, and it was suggested that these fellows might not only be going somewhere, but also doing something.
Nonetheless, there is also evidence suggesting that the mobile mRNA are just there by chance. Modelling the potential movement of the plants suggested that mobile mRNA, of which there are more than 2000, might largely be found in the phloem simply because these mRNA are hugely abundant in the companion cells. There are so many of them that, by chance, some of them get out.
This sounds like bad news for the story of mobile mRNA, but it’s not quite over yet, because a small group of mobile mRNA did not fit the model. They weren’t abundant, but they did move. And it turns out that these (and other mobile mRNAs) contain certain motifs or patterns in their code, which could potentially be acting as a signal to get going!
Even in the case of the abundant majority, it might be possible these mRNA are deliberately expressed to high levels simply in companion cells because it helps them get out into the phloem. It’s a bit of a chicken and egg situation. And it’s clear that mobile RNA have the potential to function: previous work has shown the ability for a small, interfering RNA to move from host to parasite, and ‘switch off’ a reporter gene (GUS) in that parasite!
As with many new concepts, the big picture for mRNA movement, and most importantly its mechanism and reason for existing, are still missing. But it still makes a rather fascinating tale, especially for something as ordinary as RNA.
Time to get back to the lab, eh?
References:
Spiegelman et al., (2013).: A great review on moving RNA
Calderwood et al., (2016): mRNA ability is explained by transcript abundance
Tomilov et al., (2008): Switching off a reporter gene with an siRNA
Many apologies, byt we couldn’t find an open access version of the original arabidopsis grafting paper. Please let us know in the comments if you have better luck. Here is the paywalled version: Theime et al., (2015)