Scientists, ultimately, want to understand things. And while true understanding, often comes through patience and skill – months or years of careful observation and measured experimentation – a lot of the time, the fastest way to understand something, is to F*** that something up.
‘Making mutants’ has long been the game of molecular biologists. These days, we tend to talk about genetic modification in the context of carefully considered and precisely executed manipulation of small sections of a genome – sometimes just one or two base pairs – executed with the intent to improve or ‘fix’ a certain gene and its corresponding trait. But the original genetic manipulations were strongly focused around understanding how genes work. And to understand something, sometimes the easiest thing to do is to break it.
The logic is quite simple. You break the gene, you see how the plant gets sick, and you use that sickness to understand the gene’s original function. For example, if a broken gene results in really short roots, well then, that gene might have a role in root growth. If the breaking results in pale plants, you might have yourself something involved in photosynthesis.
Apart from the easy link between ’cause’ and ‘effect, breaking genes is also relatively simple to do. In fact, many of the first ‘broken gene’ mutants were discovered in nature – plants that looked too small or too pale, and were therefore subjected to intense scientific study to work out why. All organisms occasionally acquire mutations in their DNA during their life time, but if the errors occur early enough during development, division of those cells with damaged DNA can lead to a whole organism, or sections of it, containing cells with mutations. If they occur in gamete cells – eggs or pollen – the mutations can be passed on to the next generation.

At some point, scientists realised that the natural mutation process could be sped up a little by the addition of chemicals and radiation (we’ve written about that before). These methods had the advantage of producing mutations rapidly, but blasting an entire plant’s genome tended to mean that mutations sprung up all over the genome, spread throughout multiple genes. Additionally, the mutations tended to be changes of a single letter or just a few letters in the DNA code, making them harder to find, particularly before the days of whole genome sequencing. These factors made linking an effect (a physical phenotype) to a single causative gene fairly difficult.
A little later on in the game, scientists realised that they could instead disrupt genes via exploitation of an infectious agrobacterium – Agrobacterium tumefaciens – that like to run around inserting chunks of its own DNA in the nuclear genome of the plants it infects. The large chunks, known as T-DNA, tend to insert at random positions in the nuclear genome, and if those positions happen to be in the middle of a gene, well then, that gene is broken. Because the T-DNA is large and always the same, scientists could more easily find where it landed in the genome, and thus have a fairly immediate understanding of which genes might be disrupted.
Tens of thousands of lines of Arabidopsis were created that contained T-DNA insertions in different parts of the genome, and these have subsequently been used very successfully by scientists throughout the world to understand the function of individual genes. Now, T-DNA technology is being replaced by CRISPR-based methods, which allow more specific and precise genome alterations, including deletions, insertions and small or large alterations.
While T-DNA and CRISPR techniques have catapulted plant research forward, one of the limitations of both is that they specifically target the nuclear genome of the plant. However, plants have three genomes – nuclear, plastid (chloroplast), and mitochondrial. Scientists have discovered additional reliable ways to alter the plastid genome of (some) plants. But modifying the mitochondrial genome in precise ways remains fairly unobtainable.
Which brings us back to that old-school method of deliberately making mistakes.
In order to grow and divide, cells need their DNA to replicate, and the same thing is true of the genomes of mitochondria and plastids. The machinery responsible for DNA replication, known as DNA polymerases, generally contain ‘proof reading’ capabilities, to minimise the number of mistakes made during the copying process. But the DNA polymerases can be damaged, and error-prone versions have previously been used, in the 1990s and early 2000s, as a way to induce errors into the mitochondrial genomes of yeast and mice, respectively.
In plants, replication of the mitochondrial and plastid genomes involves Plant Organellar DNA Polymerase, which are rather adorably acronym’d to ‘POP’s. POPs are encoded in the nuclear genome, made in the cytoplasm, and then imported into the mitochondria or plastid to do their job. Many plant families contain a single POP in their genomes.
The scientists chose tobacco as their study organism. This somewhat complicated things, because the tobacco nuclear genome has four copies (tetraploid) instead of the usual two (diploid), meaning it had two POPs instead of the one found in its cousin species. But working with tobacco also has some advantages (read about that here) and is a leading model species for research into the plastid and mitochondria.
The scientists identified the two tobacco POPs, and then set out to damage them, building on previous research that had successfully damaged other DNA polymerases. The scientists wanted to test whether the specific changes they were making would cause the POPs to make errors when replicating the mitochondrial genome. Ultimately, the changes could be made to the POPs within the genome of the living plants – but the process of transforming and selecting for the changes takes a relatively long amount of time. So, as a shortcut, the scientists let the damaged DNA polymerases be made in the bacteria E. coli in a test tube, and put them to work replicating a certain stretch of DNA. This DNA contained a repressor gene, which, when copied correctly and then expressed acts to prevent the expression of another gene, which in turn grants resistance to the antibiotic tetracycline. Like this:
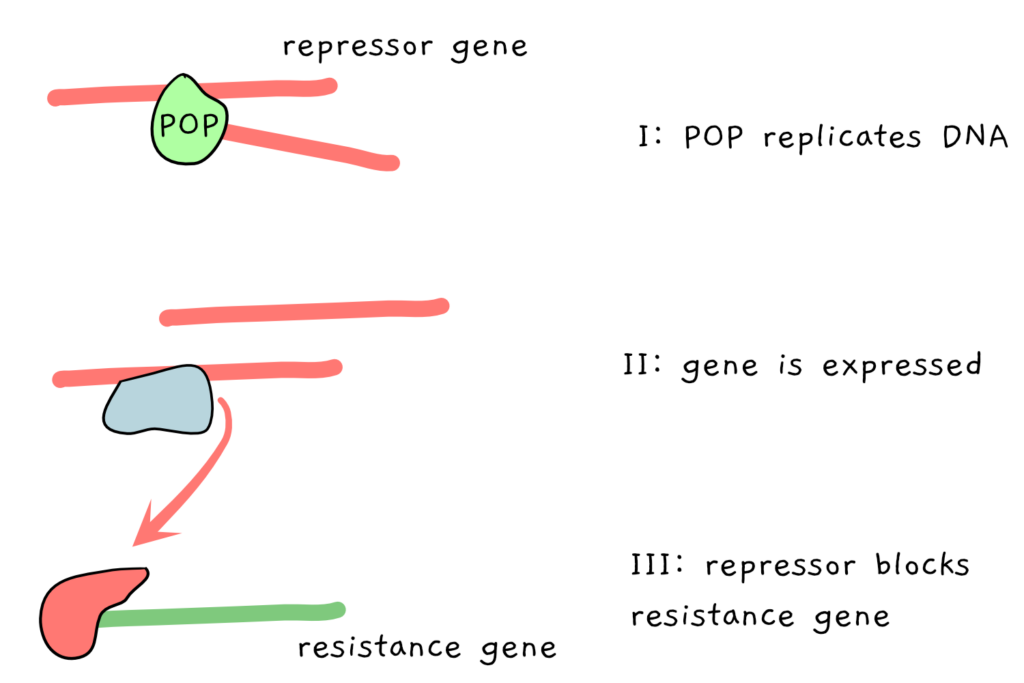
The scientists mutated the POP in various ways, looking for mutated POPs which would make more errors when copying the repressor. If the repressor is copied incorrectly, it may stop being able to do its job, and the antibiotic resistance gene ends up being expressed, instead of repressed.
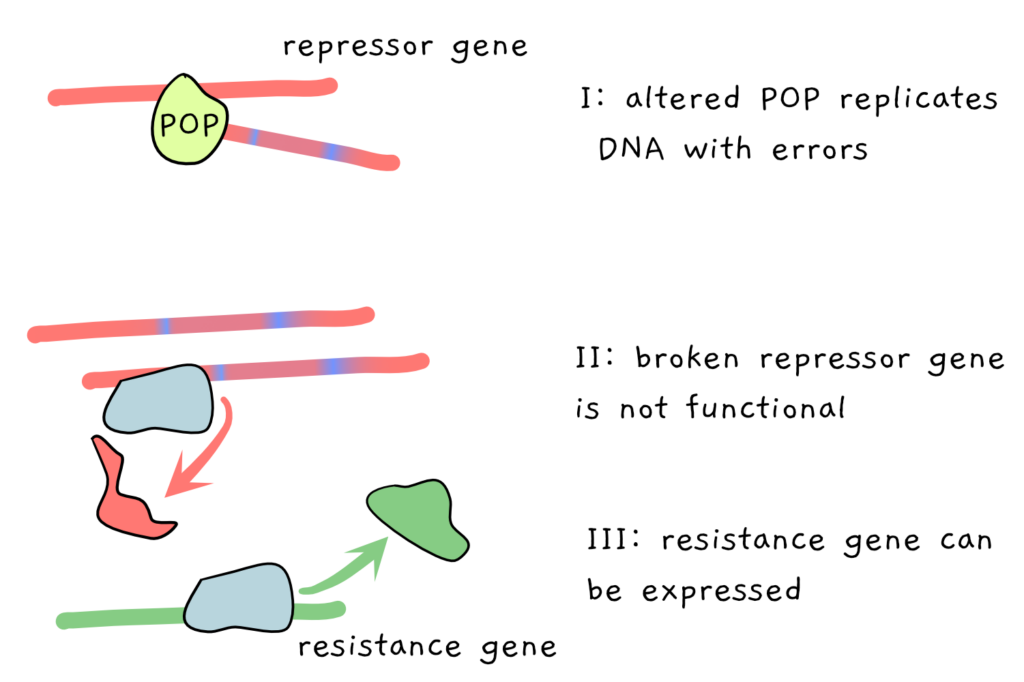
The final step is a visual readout. In this case, the E. coli containing the various POP versions is simply grown on media containing the antibiotic tetracycline. The ones with the original, unbroken POP contain functional repressors which prevent expression of tetracycline resistance: the E. coli cannot grow. But the broken POPs make mistakes as they copy, in turn producing broken repressors, which allows expression of tetracycline resistance. Suddenly, the bacteria can grow in the antibiotic!
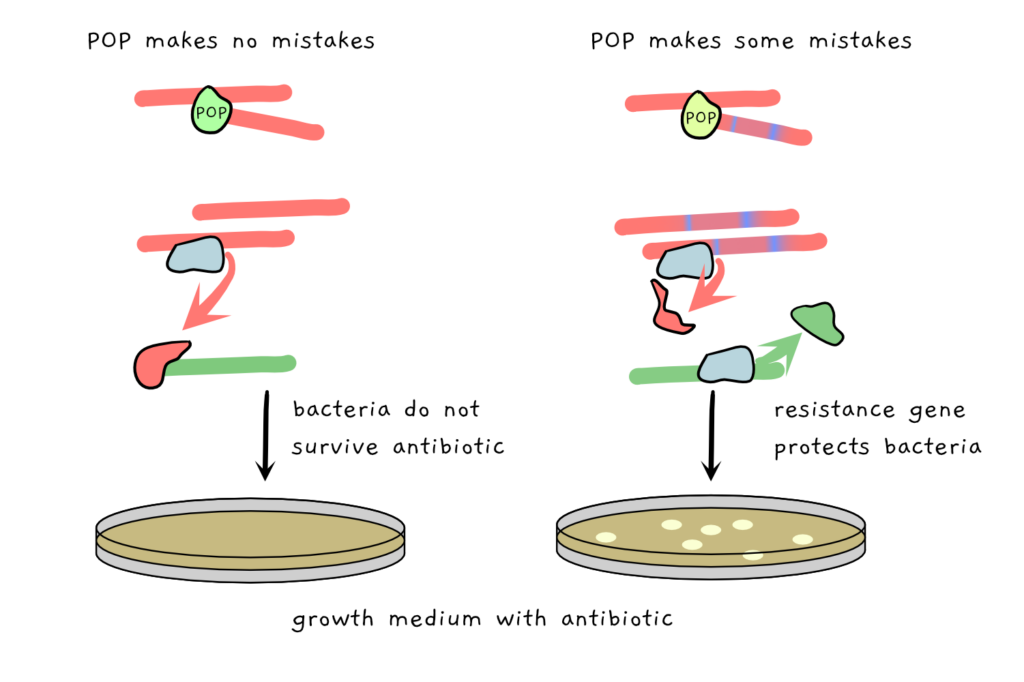
All in all, the research was successful: the scientists could make mutated POPs that were up to more than 140 times more likely to make errors when copying DNA. These errors included simple base substitutions (i.e., accidentally copying a T where an A should be), small insertions or deletions of letters, or other more complex changes.
Ultimately, this research acts as a first step. Presumably, the next stage of the research will involve mutating the tobacco POPs within the plants, and then tracking the changes made to the mitochodrial genomes. Interestingly, mitochondrial genomes are not only related to the dominant process of the mitochondria – like respiration, but have also been linked to ageing. In plants, mitochondrial genome stretches are involved in a peculiar mechanism called ‘cytoplasmic male sterility’, a biological process valuable for plant breeding. Hopefully, this new breakthrough will help further our understanding of the still-mysterious mitochondrial genome.
Reference
Junwei Ji, Anil Day, Construction of a highly error-prone DNA polymerase for developing organelle mutation systems, Nucleic Acids Research, gkaa929, https://doi.org/10.1093/nar/gkaa929