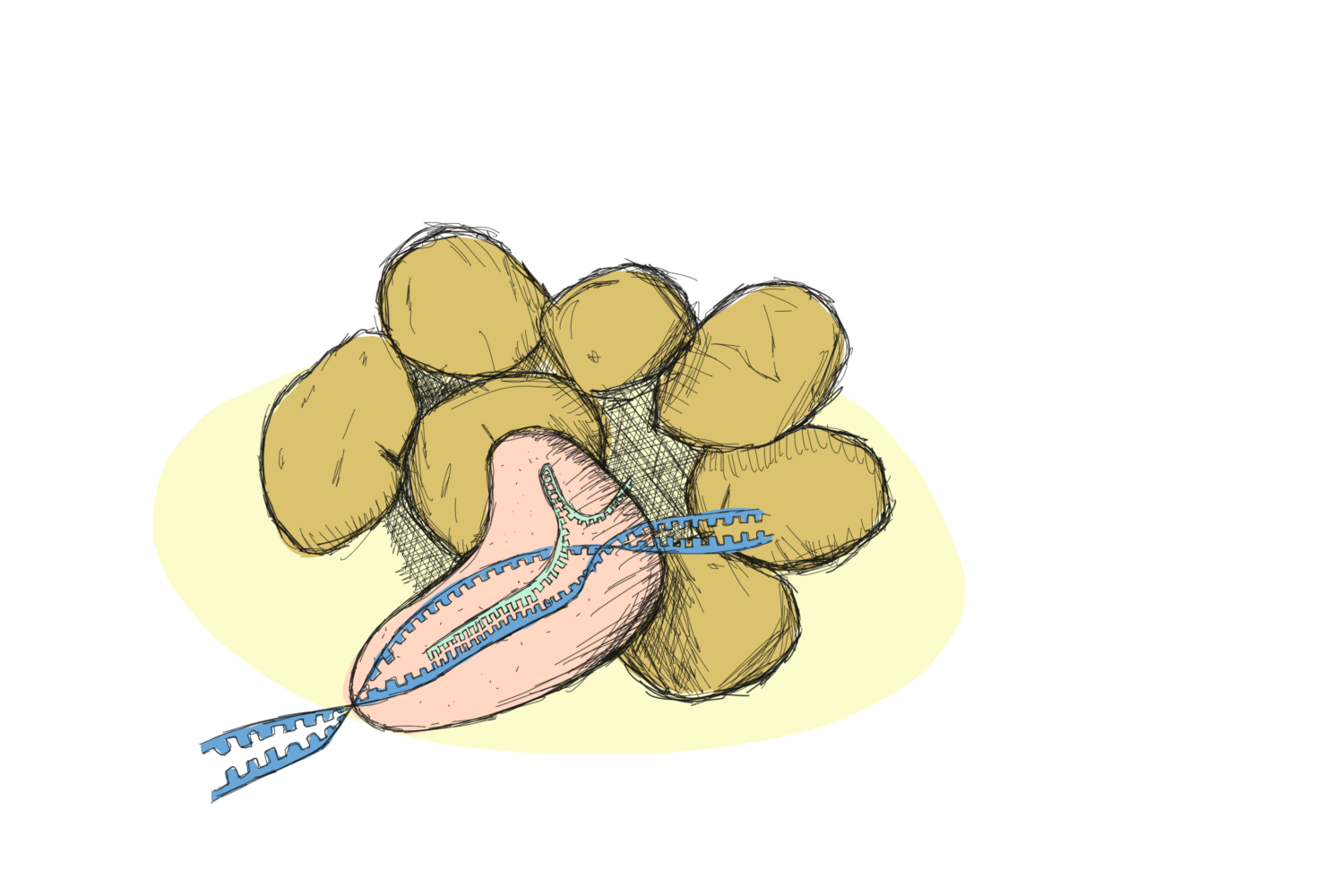
HiScientists across the globe are excited about CRISPR/Cas9 and the possibilities the new method brings to research. And they’re not alone: plant breeders are eyeballing the tool as well. What makes CRISPR/Cas9 so special in comparison to traditional breeding?
This article is part of a series on CRISPR technology. To read part one, go here.
To understand the impact of CRISPR/Cas9 on breeding, we first have to look at the traditional processes plant breeders use to create new crop varieties, or cultivars. The key factor behind new wheat, corn or lettuce varieties is genetic diversity, which is just a fancy word for small differences in the genetic code between individuals. These small differences can alter the way an organism looks or acts – for example how tall a plant grows, or how good it is at dealing with stresses, like drought or pests.
Genetic diversity is driven by mutations- usually small changes to the genetic code where one base pair (A, T, C, or G) is replaced by or changed to another, or where a base pair is lost or added. These mutations occur naturally at low rates due to reading errors by the enzymes involved in DNA replication. The rates get higher when the DNA is exposed to UV radiation (from sunlight) and rises even more when factors like nuclear radiation or specific chemicals come into play. For some 60 years, breeders have taken advantage of plant lines that were generated through mutagenesis – the process of deliberately introducing small mutations with the help of radiation or chemical agents.
Of a large pool of plants subjected to mutagenesis, some may carry mutations that are desirable – like resistance to salt stress, higher yield or a better taste. However, because the location of the changes in the genome caused by mutagenesis cannot be controlled, these plants also carry many random mutations that can have all kinds of negative effects. To get rid of all the unwanted mutations and just keep the beneficial ones, breeders perform backcrosses.
Backcrossing involves mating the mutagenised plant with an existing commercial production line, selecting the best performing offspring, and then crossing those offspring with the production line… over and over again. The aim is to dilute out all the extra mutations in the genome, keeping only those that lead to the newly found beneficial traits. Breeders must repeat this step for multiple generations, which, when generations can last for 6-8 months each, makes the process both time consuming and expensive.
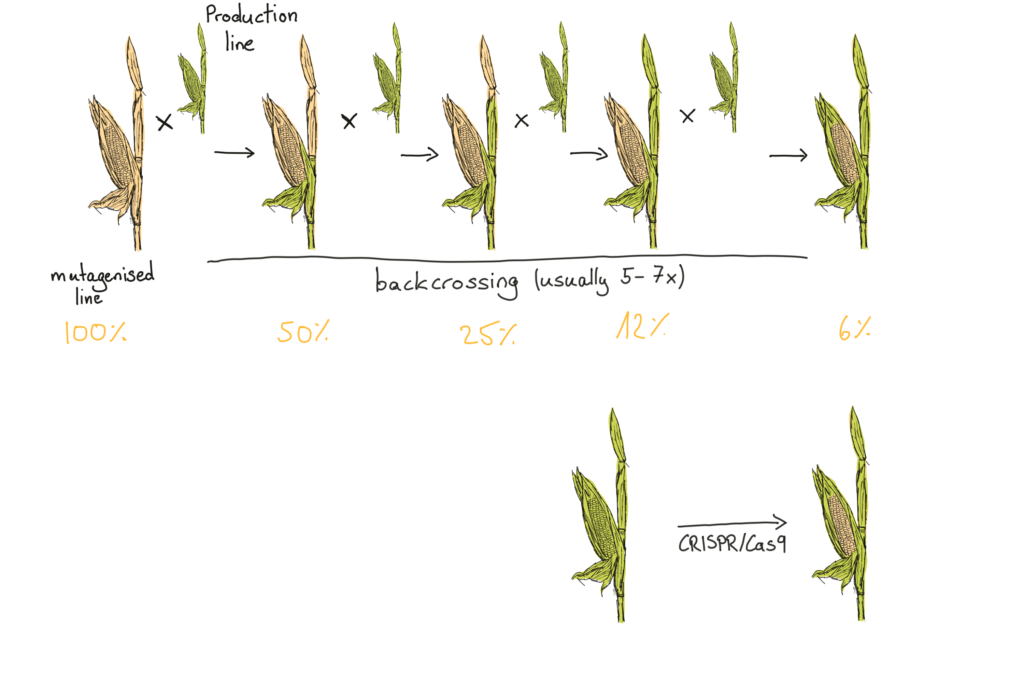
Backcrossing several times reduces the amount of mutagenised DNA in the genome to a few percent while keeping the desired trait. CRISPR/Cas9 theoretically changes nothing but the DNA linked to the desired trait.
In each generation, the ‘best performing offspring’ are selected in a process called phenotyping. Breeders measure desirable traits like yield, drought resistance, root growth behaviour or several of a dozen other factors. Measuring one or two plants is done in an afternoon, but measuring hundreds and thousand of potential candidates takes time and energy. Because the breeders cannot easily tell which of their backcrossing lines will still have the interesting trait (i.e, their mutation), and which of them will have lost it in the backcross, they have to test a whole lot of offspring. Researchers have built massive phenotyping facilities where robots autonomously move plants into measuring stations 24 hours a day and seven days a week.
Even with the help of robots, the entire process of selecting lines, crossing them back several times and testing the offspring for desirable traits takes many years and costs hundreds of thousands of Euros. Now, research is offering a shortcut – using CRISPR/Cas9.
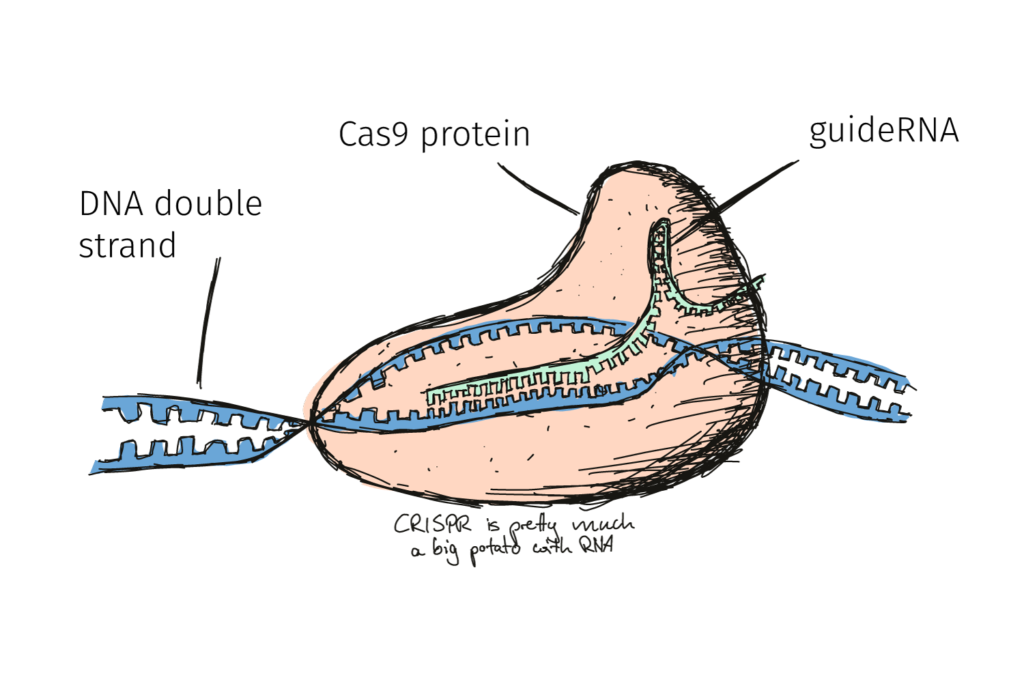
In the previous post we presented the molecular action of CRISPR/Cas9. Essentially, the method allows precise editing of the sequence of the DNA. Edits which can be very similar to a chemically-induced mutation at just the right location!
Today, breeders are able to select their gene of interest – information they take from basic plant research – edit that gene, and test for improvement. They could be done within a couple of months, using only a few plants and at a fraction of the cost.
In a fascinating experiment in 2018, researchers managed to mimic the domestication process of tomato in just one generation. They chose a wild tomato species variety and disrupted six distinct genes with CRISPR/Cas9 – in one single step. As a result, the wild, inedible tomatoes grew more like domesticated varieties and produced larger fruits. A massive shortcut – the predecessors of our commercial lines took decades to be domesticated.

The list of crops successfully edited with CRISPR/Cas9 is long. Today, we are able to use CRISPR/Cas9 in rice, wheat, maize, tomato, soybean, citrus, cotton, potato, watermelon, grapes, alfalfa, cassava, ipomoea, legumes, carrot, cacao, salvia, and lettuce. And the decreased cost of investment might make it more worthwhile to optimise more than just the biggest cash crops.
It’s not just the underdogs that could see a boost in breeding. Wheat is a cash crop with a highly complex genome containing six copies of each gene. Finding a random change in all six copies requires extensive screening and a good portion of luck. With CRISPR/Cas9, it is trivial to edit all six genes at the same time – and the final result looks like you’ve just been very lucky with your phenotyping.
Too good to be true? Well, you still can’t change everything about a plant at will. A big (and I mean BIG) prerequisite for using CRISPR/Cas9 in breeding is knowing about gene functions, their impact on plant development and their link to favorable traits. Breeders rely more than ever on the findings of basic plant research to improve their crops. Knowing the sequence of a potential target as well as the genome sequence is a major requirement. So too is having a functional transformation and regeneration system for your favourite plant. Not all crops meet these criteria – but many do.
The biggest challenge in using CRISPR/Cas9 for breeding doesn’t lie in the technology itself but in the acceptance of the technology – by society and legal bodies.
This is what we’ll talk about in the next part of this mini-series on CRISPR/Cas9: What is the public and legal opinion on genome editing?
This article is part of a mini-series on CRISPR/Cas9 in plant research. In the first part, we looked at the molecular mechanism of CRISPR/Cas9 and why it’s so exciting for researchers. In the next part, we will explore the response from society and legal bodies to CRISPR/Cas9. What is your take on the issue? Let us know in the comments!
References
European Plant Phenotyping Network, a joint effort to share phenotyping resources across Europe
Zsögön, A., Čermák, T., Naves, E. R., Notini, M. M., Edel, K. H., Weinl, S., . . . Peres, L. E. (2018, October 01). De novo domestication of wild tomato using genome editing. Nature biotechnology
Jaganathan, Deepa, Ramasamy, Karthikeyan, Sellamuthu, Gothandapani, . . . Gayatri. (2018, June 18). CRISPR for Crop Improvement: An Update Review. Frontiers in Plant Science